Long non-coding RNAs (lncRNAs) were once regarded as junk sequences. Due to the extremely complicated and inconstant mechanisms when compared with protein-coding genes, most of lncRNAs have not been well studied. CRISPR/Cas9 provides new opportunities for deeply researching lncRNA’s functions, and is receiving increasing attention in the field of lncRNA studies.
Researchers from Xishuangbanna Tropical Botanical Garden (XTBG) and Hunan Normal University constructed CRISPRlnc —a manually curated database of validated single guide RNAs (sgRNAs) for lncRNAs from all species. They got their new database published in Nucleic Acids Research.
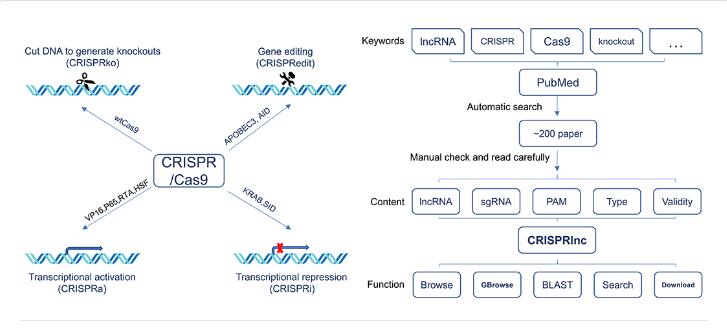
The researchers collected data upon different CRISPR/Cas9-based systems, including CRISPR knockout (CRISPRko), CRISPR activation (CRISPRa), CRISPR interference (CRISPRi) and CRISPR edit (CRISPRedit).
After manually reviewing more than 200 published papers, the new version of CRISPRlnc contains 305 lncRNAs and 2102 validated sgRNAs across eight species, including mammalian, insect and plant.
“We handled the ID, position in the genome, sequence and functional description of these lncRNAs, as well as the sequence, protoacceptor-motif (PAM) motif, CRISPR type and validity of these sgRNAs”, said Prof. LIU Changning, principal investigator of the study.
As the first database against the validated sgRNAs of lncRNAs, CRISPRlnc will provide a new and powerful approach for genome editing of lncRNAs.
“We believe that our CRISPRlnc database will thrive in the fields related to lncRNA and CRISPR/Cas9 studies, and will be popular among people in these fields”, said Prof. LIU.
For biologists to better access the information of CRISPRlnc, the researchers established a user-friendly website. Please refer to the CRISPRlnc database at http://crisprlnc.xtbg.ac.cn/ or http://www.crisprlnc.org.
Contact
LIU Channing Ph.D Principal Investigator
Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Menglun 666303, Yunnan, China
E-mail: liuchangning@xtbg.ac.cn
The CRISPRlnc database. (Image by CHEN Wen)

